Why We Age: Epigenetic Alterations
Think of DNA as the master recipe book for your body, with every cell containing a nucleus also containing a copy of this book. Interestingly, not all cells use every recipe and the same cell can change which recipes it follows over time. How is it that not all recipes are used at once in every cell, even though the book remains unchanged?
The answer to this question begins in the ancient volatile world of single-celled organisms like bacteria and archaea. While random errors in DNA replication enabled these species to diversify slowly over time, the need for more rapid adaptation to changes in temperature, pH, and other conditions necessitated additional coping strategies to ensure survival long enough to perpetuate the species [1].
As multicellular life forms evolved, these mechanisms of rapid adaptation were expanded further to capture the benefits of cellular specialization. Understanding these mechanisms that alter how DNA is expressed without changing DNA itself defines the field of epigenetics. These mechanisms act like master chefs, deciding which recipes to use and when along with how to adapt them to meet the cell’s needs and those of the greater organism [1].
Mechanisms of epigenetic regulation
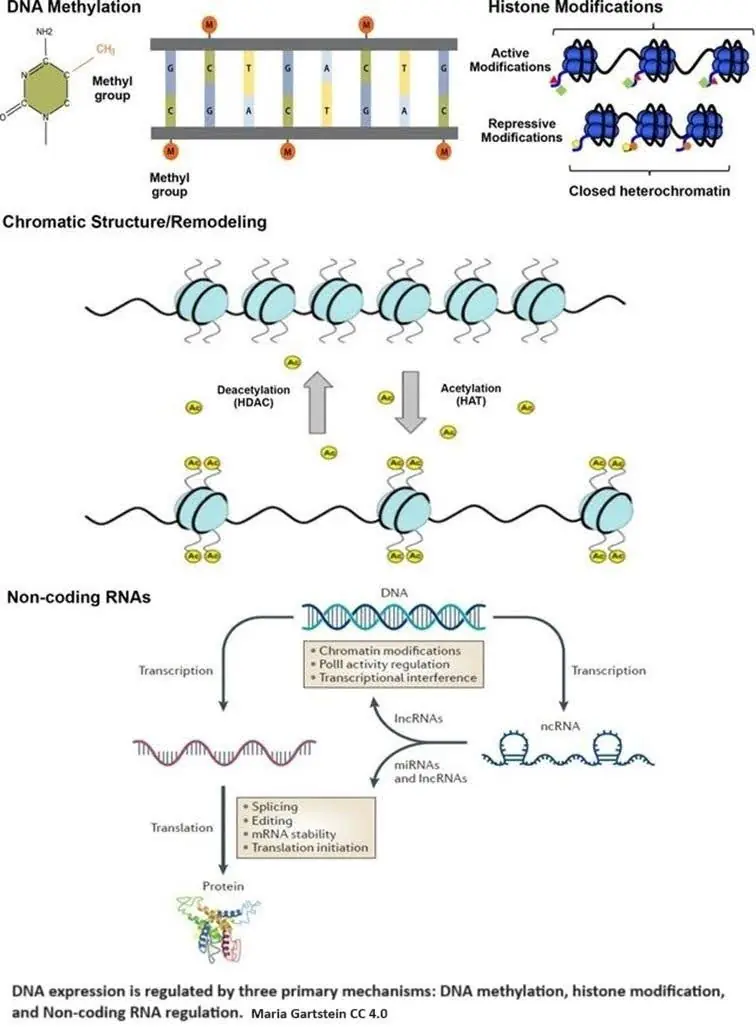
Epigenetic regulation is accomplished by three primary mechanisms: DNA methylation, histone modification, and non-coding RNA regulation.
DNA methylation occurs when a methyl group (CH3) is added to cytosine, one of the four basic building blocks of DNA. Methylation causes a cascade of effects that block transcription machinery and cause chromatin to condense.
If DNA is the recipe book, transcription is copying a recipe from the book to deliver to the chef. Chromatin is made up of DNA and histone proteins, the tiny molecular spools around which DNA is wrapped. This both organizes DNA and determines what DNA recipes are available. DNA wrapped around the spools is essentially hidden. Condensation means packing the DNA together tightly by spooling it up.
Histone modifications, such as acetylation and methylation, are crucial in regulating how tightly DNA is wound around histones, exposing or hiding specific genes from the cell’s machinery. In epigenetics, acetylation primarily involves adding an acetyl group (CH3CO) to the amino acid lysine.
Why lysine? It has a long tail that sticks out of the histone and carries a positive charge. DNA typically carries a negative charge; since negative and positive charges are attracted to each other, this keeps the DNA and the histones spooled up. The positive charge disappears by adding the acetyl group, and DNA unwinds from the histone, exposing the recipes. Histone acetyltransferases (HATs) are enzymes that help the acetyl group attach to lysine.
Non-coding RNAs either modify histones or attach themselves to mRNA transcripts, which blocks them from being translated into proteins or leads to their degradation [2]. Metaphorically, non-coding RNAs intercept the recipe copy and either redact part of it or throw it in the garbage.
Like a symphony, these epigenetic mechanisms work harmoniously to ensure that the right genes are expressed at the right time and place, allowing for the specialization of cell functions. The maintenance of these epigenetic marks is a delicate dance. As this dance is disrupted, the cell’s identity and ability to fulfill its mission are significantly altered.
Epigenetic alterations and aging
Epigenetic alterations refer to the gradual loss of cellular identity and are considered a primary hallmark of aging in López-Otín and Blasco’s seminal work “The Hallmarks of Aging.” The primary hallmarks of aging are significant because they drive many “downstream” aging processes [3].
Epigenetic alterations lead to the dysregulation of gene expression, thereby impacting cellular function and identity. This dysregulation results in genomic instability, another hallmark of aging, as the epigenetic machinery involved in maintaining DNA integrity becomes compromised. Additionally, altered epigenetic states can lead to cellular senescence, which promotes inflammation and tissue dysfunction [3].
Epigenetic alterations also affect mitochondrial function and bioenergetics by modulating the expression of genes involved in mitochondrial maintenance and energy production, contributing to the decline in cellular metabolism observed with aging. Furthermore, impaired epigenetic regulation interferes with proteostasis by altering the expression of chaperones and proteolytic systems, accumulating damaged proteins [4].
Lastly, the chronic inflammation seen in aging, known as “inflammaging,” is often driven by epigenetic alterations that result in the persistent activation of pro-inflammatory genes. Thus, understanding the nature of epigenetic regulatory systems and epigenetic alterations is essential. They underpin many biological processes that drive aging, making them a key target for interventions to promote healthy aging and mitigate age-related diseases [5].
This knowledge enables the development of novel therapeutics that can restore healthy gene function. It paves the way for personalized medicine approaches that tailor treatments based on an individual’s unique epigenetic landscape, ultimately improving health outcomes and extending healthy lifespan.
Principles of epigenetic reprogramming
Epigenetic reprogramming refers to resetting the epigenetic marks in a cell to a more youthful state. This involves altering DNA methylation, histone modifications, and non-coding RNA expression to restore the gene expression profile associated with a younger appearance or phenotype [6].
The primary goal is to remove age-associated epigenetic changes. By reprogramming their epigenetic marks, cells can regain their youthful function and regenerative capacity. This can be achieved through various techniques, including specific reprogramming factors, small molecules, and advanced gene-editing technologies like CRISPR [7, 8].
Key studies and potential for reversing aging processes
The discovery of the Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc: OSKM) has been a significant breakthrough in epigenetic reprogramming. These factors can reprogram somatic cells into induced pluripotent stem cells (iPSCs), effectively resetting their epigenetic age. In a landmark study by Takahashi and Yamanaka (2006), introducing these factors into adult cells resulted in their reversion to a pluripotent state, demonstrating the feasibility of epigenetic reprogramming [9].
Partial reprogramming involves temporarily expressing reprogramming factors to rejuvenate cells without fully reverting them to a pluripotent state, thus reducing the risk of tumor formation. Ocampo et al. (2016) demonstrated that cyclic partial reprogramming in a mouse model could extend lifespan and improve tissue regeneration without causing tumors, showing potential for reversing age-related changes in a controlled manner [10].
CRISPR technology has been adapted to target and modify specific epigenetic marks without altering the underlying DNA sequence. This allows for precise control over gene expression and restoring youthful gene expression patterns. A recent study by Park et al. (2022) used CRISPR/Cas9 to target and demethylate specific genes associated with aging, resulting in improved cellular function and longevity in a mouse model [11].
Small molecules modulating epigenetic enzymes, such as DNMT and HDAC inhibitors, are being explored for their potential to reverse aging. These compounds can alter DNA methylation and histone modification patterns to rejuvenate cells. For example, DNMT inhibitors like 5-azacytidine have shown promise in rejuvenating aged cells by demethylating age-related hypermethylated genes and restoring their expression [12].
Combining senolytic drugs that selectively eliminate senescent cells with epigenetic techniques can enhance rejuvenation. This approach aims to improve overall tissue function by clearing senescent cells and reprogramming the remaining healthy cells. A study by Zhu et al. (2015) demonstrated that senolytic treatment, combined with siRNA to knock down pro-survival genes, improved tissue function and extended lifespan in aged mice [13].
Small interfering RNAs (siRNAs) were used to silence the expression of key nodes of anti-apoptotic networks, such as EFNB1, EFNB3, PI3KCD, and p21, selectively killing senescent cells without affecting non-senescent cells. siRNA, as a form of non-coding RNA, plays a role in the epigenetic regulation of DNA expression, contributing to the reprogramming and rejuvenation of cells [13].
Epigenetic clocks, which measure biological age based on DNA methylation patterns, allow researchers to monitor the effectiveness of reprogramming interventions. Horvath’s epigenetic clock (2013) has been widely used to assess the biological age of cells and tissues, providing a valuable tool for evaluating the success of reprogramming efforts [14].
Epigenetic reprogramming holds significant potential for combating aging by restoring youthful gene expression patterns and cellular function. Key studies have demonstrated the feasibility of reprogramming cells to a more youthful state using Yamanaka factors, CRISPR-based epigenetic editing, small molecules, and combination therapies [9, 11]. Ongoing research in this field aims to refine these techniques and develop safe, effective interventions for reversing age-related changes and extending healthy lifespan.
Lifestyle and epigenetics
Lifestyle changes, particularly regular physical activity, have been shown to induce beneficial epigenetic changes. Exercise can lead to changes in DNA methylation patterns, particularly in genes involved in energy metabolism, muscle growth, and anti-inflammatory pathways, thereby enhancing their expression. Exercise also influences histone acetylation and methylation, resulting in more relaxed chromatin structures and making genes more accessible for transcription [15].
Furthermore, physical activity can alter the expression of miRNAs and lncRNAs, which play roles in muscle adaptation, inflammation regulation, and overall cellular stress response. Exercise demethylates the promoter region of the PGC-1α gene, a key regulator of mitochondrial biogenesis and oxidative metabolism, enhancing its expression [16]. Similarly, genes like NRF1 and TFAM, involved in mitochondrial function, show altered methylation and increased expression in response to exercise [17].
Research on identical twins has shown that people who engage in regular physical activity have distinct DNA methylation patterns compared to their sedentary counterparts, particularly in genes associated with inflammation and metabolism. Long-term studies have demonstrated that people who maintain regular physical activity throughout their lives have epigenetic profiles associated with better metabolic health, reduced inflammation, and lower risks of age-related diseases [18].
In rodent studies, exercise interventions have shown significant changes in DNA methylation and histone modification in muscle, liver, and brain tissues, correlating with improved metabolic function, neurogenesis, and cognitive function. Older rodents subjected to exercise regimens exhibit epigenetic changes that mitigate age-related declines in muscle function and cognitive abilities [19].
Chronic stress can lead to hypermethylation or hypomethylation of specific genes, particularly those involved in the stress response, such as NR3C1, the glucocorticoid receptor gene, altering their expression and affecting how the body responds to stress [20]. Stress can also alter histone acetylation and methylation patterns, leading to changes in chromatin structure and gene expression, often resulting in the repression of genes involved in neuroprotection and the activation of genes that promote inflammation.
Stress influences the expression of miRNAs and lncRNAs that regulate genes involved in inflammation, synaptic plasticity, and stress responses. For example, hypermethylation of the NR3C1 gene promoter in individuals exposed to chronic stress is associated with reduced expression of glucocorticoid receptors, impairing the body’s ability to regulate the stress response [21]. Reduced expression of brain-derived neurotrophic factor (BDNF) due to altered histone acetylation and methylation is linked to stress-related psychiatric disorders, such as depression and anxiety.
Stress reduction techniques, such as mindfulness and meditation, have been shown to reduce stress-induced epigenetic changes [20]. Practices like mindfulness and meditation can lead to the demethylation of genes involved in the stress response and the acetylation of histones associated with relaxation and neuroplasticity.
Participants in mindfulness-based stress reduction (MBSR) programs exhibit changes in DNA methylation patterns in genes related to immune function and stress response, correlating with reduced stress and improved well-being [22]. Individuals practicing transcendental meditation show decreased methylation in stress-related genes and altered expression of miRNAs involved in inflammation regulation.
Regular practice of yoga has been associated with changes in DNA methylation and histone modifications that enhance the expression of anti-inflammatory genes and reduce stress-related gene activation. Techniques such as deep breathing and progressive muscle relaxation can induce epigenetic changes that improve immune function and reduce the expression of pro-inflammatory genes [23, 24].
Exercise not only induces beneficial epigenetic changes on its own but also helps mitigate the epigenetic impacts of stress. Regular physical activity can normalize stress-induced epigenetic modifications, promoting resilience and overall health [25].
Lifestyle changes, including regular physical activity and effective stress management techniques, have profound impacts on the epigenetic landscape. Exercise-induced epigenetic changes enhance metabolic health, muscle function, and inflammation regulation.
Stress reduction techniques can also counteract the detrimental epigenetic effects of chronic stress, promoting better mental health and resilience. Lifestyle interventions, therefore, are critical in maintaining and improving health through epigenetic regulation [26].
Dietary habits shape the epigenetic landscape by taking specific nutrients and bioactive compounds. Nutrients such as folate, B vitamins, and choline are essential for maintaining proper DNA methylation [27].
Folate, or Vitamin B9, is vital in the one-carbon metabolism pathway, which provides methyl groups for DNA methylation. It is converted to tetrahydrofolate (THF), which participates in synthesizing S-adenosylmethionine (SAM), the primary methyl donor in cellular methylation reactions.
Adequate folate levels are essential for maintaining proper DNA methylation patterns, as deficiency can lead to global DNA hypomethylation, associated with genomic instability and increased cancer risk.
Vitamin B12, or cobalamin, works with folate in the one-carbon metabolism pathway and is a cofactor for methionine synthase, which converts homocysteine to methionine, subsequently forming SAM [28]. Adequate levels of Vitamin B12 support proper DNA methylation and gene regulation, while deficiency can disrupt methylation patterns and potential health issues, including neurological disorders.
Vitamin B6, or pyridoxine, is involved in the conversion of homocysteine to cysteine, indirectly supporting the methylation cycle by preventing the accumulation of homocysteine. By maintaining the balance of one-carbon metabolism, Vitamin B6 ensures the availability of methyl groups for DNA methylation.
Choline, a precursor for betaine, acts as a methyl donor in the conversion of homocysteine to methionine, supporting SAM synthesis and proper methylation processes and gene regulation [27].
Dietary sources of these nutrients include leafy green vegetables, legumes, nuts, and fortified grains for folate; animal products and fortified cereals for Vitamin B12; poultry, fish, potatoes, chickpeas, and bananas for Vitamin B6; and eggs, liver, fish, nuts, and cruciferous vegetables for choline.
Folate supplementation can correct DNA hypomethylation, reduce the risk of neural tube defects in pregnancy, and lower the risk of colorectal cancer. Vitamin B12 supplementation has been shown to improve DNA methylation status and potentially mitigate cognitive decline in the elderly.
Supplementation with B6 and choline has been associated with improved methylation patterns and reduced levels of homocysteine, a marker for cardiovascular disease [29].
Bioactive compounds such as polyphenols also play a significant role in modulating histone modifications. Polyphenols, found in plants, have antioxidant and anti-inflammatory properties and can modulate histone modifications, such as acetylation and methylation, influencing chromatin structure and gene expression.
Resveratrol, found in red grapes, red wine, berries, and peanuts, activates sirtuins, particularly SIRT1, a class of histone deacetylases (HDACs) that remove acetyl groups from histones, leading to chromatin condensation and altered gene expression [30]. Resveratrol influences histone acetylation and methylation, promoting the expression of genes involved in longevity, stress resistance, and metabolic regulation.
Curcumin, derived from the turmeric root, can inhibit HDACs and histone acetyltransferases (HATs), modulating histone acetylation and affecting chromatin structure and gene expression. Curcumin alters histone acetylation and methylation patterns, leading to the activation of tumor suppressor genes and the inhibition of inflammatory pathways.
Studies on dietary interventions have demonstrated the potential of these compounds to influence gene expression and improve health outcomes. Human studies have shown that resveratrol supplementation in obese men led to increased SIRT1 activity, improved insulin sensitivity, and altered the expression of genes involved in glucose metabolism, with beneficial metabolic effects and potential anti-aging benefits [30].
Animal studies have shown that resveratrol administration can extend lifespan and improve healthspan by modulating epigenetic marks associated with aging and metabolic regulation. Resveratrol-treated animals exhibited improved glucose tolerance, reduced inflammation, and enhanced mitochondrial function [31].
Research in patients with colorectal cancer demonstrated that curcumin supplementation resulted in the reactivation of silenced tumor suppressor genes through changes in histone acetylation, correlating with reduced tumor growth and improved clinical outcomes [32].
In rodent models of neurodegenerative diseases, curcumin supplementation improved cognitive function and reduced neuroinflammation by modulating histone acetylation and methylation, with neuroprotective effects and potential therapeutic benefits [33].
In conclusion, dietary habits significantly influence the epigenetic landscape through the intake of specific nutrients and bioactive compounds. Nutrients such as folate, B vitamins, and choline are crucial for maintaining proper DNA methylation, while polyphenols like resveratrol and curcumin can modulate histone modifications.
Studies on dietary interventions have demonstrated the potential of these compounds to influence gene expression, improve metabolic health, and provide anti-aging benefits. This underscores the importance of nutritional strategies in promoting health and preventing disease through epigenetic regulation.
The gut microbiome
The gut microbiome, consisting of trillions of microorganisms, including bacteria, viruses, fungi, and other microbes residing in the gastrointestinal tract, plays crucial roles in digestion, immune function, and overall health [34].
These microorganisms can influence epigenetic mechanisms such as DNA methylation, histone modifications, and non-coding RNA activity, affecting gene expression and cellular function [35]. The gut microbiome produces various metabolites, such as short-chain fatty acids (SCFAs), vitamins, and bile acids, which can modulate epigenetic marks.
Additionally, the gut microbiome interacts with the immune system, influencing inflammation and immune responses that can affect epigenetic regulation. Furthermore, the microbiome communicates with the brain through the gut-brain axis, impacting neurological function and behavior through epigenetic mechanisms.
Understanding the intricate relationship between the gut microbiome and epigenetic regulation provides insights into how microbial communities within the gut can influence overall health and disease processes [36-38].
Epigenetic drugs
Epigenetic regulation is crucial in controlling gene expression and maintaining cellular function. Disruptions in epigenetic mechanisms are implicated in various diseases, including cancer, neurodegenerative disorders, and metabolic conditions.
Epigenetic drugs target these regulatory mechanisms to restore normal gene function and treat disease. The main classes of epigenetic drugs include DNA methyltransferase (DNMT) inhibitors, histone deacetylase (HDAC) inhibitors, histone methyltransferase (HMT) inhibitors, bromodomain and extra-terminal (BET) inhibitors, lysine-specific demethylase 1 (LSD1) inhibitors, and CRISPR-based epigenetic editing technologies [39, 40].
DNA methyltransferase (DNMT) inhibitors
DNMT inhibitors alter the epigenetic landscape by targeting DNA methylation patterns. These inhibitors have shown promise in treating various cancers and potentially reversing aging by reactivating silenced genes [41].
Azacitidine (Vidaza)
Azacitidine, marketed as Vidaza, is a nucleoside analog that incorporates into DNA and RNA, inhibiting DNMTs. A nucleoside analog looks like a component of DNA but is a little different. This inhibition leads to DNA hypomethylation and subsequent reactivation of silenced genes, including tumor suppressor genes.
Azacitidine was first approved by the FDA in 2004 for the treatment of myelodysplastic syndromes (MDS), a group of hematologic cancers characterized by ineffective blood cell production and a high risk of progression to acute myeloid leukemia (AML).
The drug’s primary benefit lies in its ability to reduce the need for blood transfusions and delay the progression to leukemia. However, azacitidine also presents risks, including a reduction in blood cells (cytopenia), gastrointestinal disturbances, and an increased risk of infections due to its non-specific hypomethylation effects [42, 43].
Decitabine (Dacogen)
Decitabine, marketed as Dacogen, is another DNMT inhibitor that irreversibly binds to DNMTs, leading to DNA hypomethylation and reactivation of silenced genes. Approved by the FDA in 2006 for the treatment of myelodysplastic syndromes (MDS) and later for acute myeloid leukemia (AML) in older patients or those ineligible for standard chemotherapy.
Decitabine works similarly to azacitidine by incorporating into DNA and causing hypomethylation. Decitabine’s benefits include improved overall survival rates and reduced disease progression. However, like azacitidine, decitabine carries significant risks, such as myelosuppression, increased susceptibility to infections, and gastrointestinal issues [44].
Recent developments
Clinical trials have explored the expanded use of DNMT inhibitors like azacitidine and decitabine in treating other cancers beyond MDS and AML. Studies have investigated their efficacy in solid tumors and additional hematologic malignancies.
For instance, trials combining DNMT inhibitors with immune checkpoint inhibitors or targeted therapies have shown promising results, enhancing the therapeutic effects and potentially overcoming resistance to conventional treatments [45].
A notable 2024 study identified specific DNA methylation biomarkers that predict the response to DNMT inhibitors in MDS patients, improving treatment stratification and outcomes [46]. This advancement underscores the potential for more personalized approaches in using DNMT inhibitors.
Additionally, the combination of DNMT inhibitors with other therapeutic agents is an area of active research. Combining these inhibitors with drugs targeting different epigenetic or signaling pathways aims to enhance their efficacy and reduce resistance.
Recent trials have demonstrated that such combination therapies can lead to improved patient outcomes, particularly in cancers that are refractory to single-agent treatments [47].
In summary, the development and use of DNMT inhibitors, such as azacitidine and decitabine, have significantly impacted the treatment of MDS and AML. Ongoing research and clinical trials continue to expand their applications, aiming to improve efficacy and patient outcomes through combination therapies and personalized treatment strategies. While these drugs offer substantial benefits, their use must be carefully managed to mitigate the associated risks.
Histone deacetylase (HDAC) inhibitors
Histone deacetylase (HDAC) inhibitors are a class of drugs that modulate the acetylation status of histones, leading to changes in chromatin structure and gene expression.
These inhibitors have shown efficacy in treating various cancers by reactivating silenced tumor suppressor genes and altering the transcriptional landscape of cancer cells. More recent studies suggest this class of drugs may have potential in the treatment of Alzheimer’s disease [48].
Vorinostat (SAHA)
Vorinostat, also known as suberoylanilide hydroxamic acid (SAHA), was the first HDAC inhibitor approved by the FDA in 2006. It works by inhibiting histone deacetylases and accumulating acetylated histones, which results in a more relaxed chromatin structure and increased gene expression.
Vorinostat is approved for the treatment of cutaneous T-cell lymphoma (CTCL), particularly in patients with progressive, persistent, or recurrent disease. The benefits of vorinostat include inducing apoptosis and cell cycle arrest in cancer cells, thereby reducing tumor growth.
However, the drug can cause several side effects, including fatigue, nausea, vomiting, diarrhea, thrombocytopenia, and potential cardiac arrhythmias [49].
Romidepsin (Istodax)
Romidepsin, marketed as Istodax, is a cyclic peptide HDAC inhibitor that was approved by the FDA in 2009 for the treatment of CTCL and peripheral T-cell lymphoma (PTCL).
Romidepsin functions by inhibiting HDAC enzymes, leading to increased acetylation of histones, and promoting the expression of genes involved in cell cycle regulation and apoptosis.
The clinical benefits of romidepsin include its ability to induce tumor cell death and improve patient survival rates. However, it also presents risks, such as nausea, fatigue, infections due to myelosuppression, and electrolyte imbalances [50].
Belinostat (Beleodaq)
Belinostat, sold under the brand name Beleodaq, is a pan-HDAC inhibitor approved by the FDA in 2014 for the treatment of peripheral T-cell lymphoma (PTCL). Belinostat inhibits multiple HDAC enzymes, resulting in increased acetylation of histones and non-histone proteins, which leads to the activation of tumor suppressor genes and apoptosis in cancer cells.
The approval of belinostat was based on its demonstrated efficacy in inducing responses in patients with PTCL, including those who had previously received other treatments.
Benefits of belinostat include its potential to improve overall survival and reduce tumor burden. However, it can cause adverse effects such as anemia, thrombocytopenia, nausea, vomiting, and liver toxicity [51, 52].
Recent developments
The development and clinical use of HDAC inhibitors have continued to evolve, with research focusing on improved formulations, expanded indications, and combination therapies. Newer HDAC inhibitors are being designed to enhance pharmacokinetics and reduce side effects [53]. Additionally, HDAC inhibitors are being tested in combination with other anticancer agents, such as immune checkpoint inhibitors, targeted therapies, and chemotherapeutics, to enhance their efficacy and overcome resistance mechanisms [45].
Clinical trials have shown promising results, particularly in solid tumors and neurodegenerative diseases [48], highlighting the potential of HDAC inhibitors beyond hematologic malignancies.
In conclusion, HDAC inhibitors like vorinostat, romidepsin, and belinostat have become essential tools in treating certain lymphomas. Their ability to alter the epigenetic landscape of cancer cells provides a mechanism to reactivate tumor suppressor genes and induce apoptosis.
Ongoing research and clinical trials continue to expand their therapeutic potential, aiming to improve outcomes for patients with various cancers and other diseases.
HDAC6 inhibitors
Histone deacetylase 6 (HDAC6) inhibitors specifically target HDAC6, an enzyme that deacetylates non-histone proteins, including α-tubulin and heat shock protein 90 (HSP90). This enzyme is crucial in cellular processes like protein degradation and stress response [54].
Ricolinostat (ACY-1215)
Ricolinostat, or ACY-1215, was initially considered a selective HDAC6 inhibitor. By inhibiting HDAC6, ricolinostat increases acetylation of α-tubulin, resulting in disrupted microtubule function and enhanced protein degradation through the aggresome pathway.
This disruption induces apoptosis in cancer cells. Ricolinostat has been investigated primarily for its potential in treating multiple myeloma [55]. Clinical trials have shown that when combined with proteasome inhibitors such as bortezomib, ricolinostat enhances the efficacy of these treatments [56].
The benefits of ricolinostat include its ability to induce cancer cell death and enhance the effectiveness of existing therapies. However, risks include gastrointestinal symptoms, fatigue, and hematological toxicity, necessitating careful management of dosing.
Tucidinostat (HBI-8000)
Tucidinostat, also known as HBI-8000, is a selective HDAC6 inhibitor that has shown promise in treating various hematologic malignancies and solid tumors. Tucidinostat works by inhibiting HDAC6, accumulating acetylated proteins, which disrupts cellular processes critical for cancer cell survival.
Tucidinostat was approved in China for the treatment of peripheral T-cell lymphoma (PTCL) in 2014. Clinical studies have demonstrated its efficacy in inducing tumor regression and improving patient outcomes.
This drug can be potentially used in combination with other anticancer agents to enhance therapeutic effects. However, it can cause side effects such as fatigue, gastrointestinal disturbances, and hematological toxicity [57].
Recent developments in HDAC6 inhibitors
Recent research has focused on improving the formulations of HDAC6 inhibitors to enhance their pharmacokinetic properties and reduce side effects [58]. Additionally, studies are expanding the use of these inhibitors to treat solid tumors and neurodegenerative diseases.
For instance, HDAC6 inhibitors have shown potential in improving neurodegenerative conditions by modulating protein degradation pathways and reducing neuroinflammation [59].
Clinical trials continue to explore the efficacy of HDAC6 inhibitors in various cancers, with promising results indicating their potential to enhance existing therapies and provide new treatment options.
Histone methyltransferase (HMT) inhibitors
Histone methyltransferase inhibitors target enzymes that add methyl groups to histones, thereby influencing gene expression. EZH2 inhibitors are a notable class within this category [60].
EZH2 inhibitors: Tazemetostat (Tazverik)
Tazemetostat, marketed as Tazverik, is an EZH2 inhibitor approved by the FDA in 2020 for treating epithelioid sarcoma and follicular lymphoma. EZH2 is a histone methyltransferase that catalyzes the addition of methyl groups to histone H3 at lysine 27 (H3K27), leading to gene silencing.
Tazemetostat inhibits EZH2, reducing this methylation and reactivating silenced genes, including tumor suppressor genes. The benefits of Tazemetostat include its ability to inhibit cancer cell proliferation and induce tumor regression.
However, risks include fatigue, nausea, thrombocytopenia, and potential secondary malignancies due to non-specific inhibition of histone methylation [61-63].
Recent developments in EZH2 inhibitors
Research on EZH2 inhibitors has expanded to explore their use in combination therapies. Combining EZH2 inhibitors with other treatments, such as immune checkpoint inhibitors and targeted therapies, has shown enhanced efficacy in preclinical and clinical studies.
Clinical trials are investigating these combinations in various cancer types, aiming to improve patient outcomes by leveraging the synergistic effects of multiple therapeutic agents [64, 65].
Bromodomain and extra-terminal (BET) inhibitors
BET inhibitors target bromodomain proteins that recognize acetylated lysines on histones, playing a crucial role in regulating gene expression by recruiting transcriptional machinery [66].
JQ1
JQ1 is a pioneering BET inhibitor that disrupts the interaction between BET proteins and acetylated histones, thereby decreasing the expression of oncogenes and other pathogenic genes.
JQ1 has shown significant promise in preclinical studies for various cancers, including leukemia and solid tumors. The benefits of JQ1 include its ability to suppress oncogene expression, induce cell cycle arrest, and promote apoptosis in cancer cells.
However, its clinical development has been limited by challenges such as potential resistance mechanisms and side effects like gastrointestinal disturbances and thrombocytopenia [67].
I-BET762
I-BET762, also known as Molibresib, is another BET inhibitor that has demonstrated anti-tumor activity by modulating the expression of key regulatory genes in cancer cells. It works similarly to JQ1 by disrupting the binding of BET proteins to acetylated histones, leading to altered gene expression.
I-BET762 has been evaluated in clinical trials for various cancers, including lymphoma and solid tumors. The benefits of I-BET762 include its potential to suppress the expression of oncogenes and induce apoptosis.
However, it also carries risks, including liver toxicity, gastrointestinal symptoms, and the development of resistance, necessitating the exploration of combination therapies [68].
Recent developments in BET inhibitors
BET inhibitors are being investigated as treatments against various cancers, particularly in combination therapies to enhance their efficacy. Clinical trials are exploring the combination of BET inhibitors with other anticancer agents, such as immune checkpoint inhibitors and targeted therapies, to overcome resistance and improve patient outcomes.
These combination approaches have shown promising results in preclinical studies and early-phase clinical trials, indicating the potential for BET inhibitors to become integral components of multi-modal cancer therapy [68].
The development and clinical application of HDAC, EZH2, and BET inhibitors represent significant advancements in the field of epigenetic therapies. By targeting specific epigenetic mechanisms, these drugs have shown efficacy in treating various cancers and hold promise for expanding indications and combination strategies.
Ongoing research and clinical trials continue to refine these therapies, aiming to maximize their therapeutic potential while minimizing adverse effects. As our understanding of epigenetic regulation deepens, these inhibitors will likely play an increasingly vital role in personalized medicine and the treatment of complex diseases.
Lysine-specific demethylase 1 (LSD1) inhibitors
Lysine-specific demethylase 1 (LSD1) inhibitors target the LSD1 enzyme, which demethylates histone H3 on lysine 4 (H3K4) and lysine 9 (H3K9), thereby regulating gene expression. Inhibition of LSD1 can lead to the reactivation of genes involved in differentiation and apoptosis, making it a promising target for cancer therapy [69].
ORY-1001
ORY-1001 is a potent and selective LSD1 inhibitor that has shown promise in treating acute myeloid leukemia (AML) and other hematologic malignancies. The mechanism of action involves inhibiting LSD1, leading to the accumulation of methylated histones and reactivation of silenced genes that promote differentiation and apoptosis of cancer cells.
Clinical trials have demonstrated that ORY-1001 can effectively reduce leukemic blasts and improve patient outcomes. The benefits of ORY-1001 include its ability to induce differentiation and apoptosis in cancer cells, potentially leading to durable remissions.
However, risks include hematological toxicity, fatigue, and gastrointestinal disturbances, which require careful monitoring and management [70].
IMG-7289 (Bomedemstat)
IMG-7289, also known as Bomedemstat, is another LSD1 inhibitor that targets the demethylase activity of LSD1, leading to the reactivation of silenced genes involved in cell differentiation and apoptosis.
IMG-7289 has shown efficacy in preclinical and clinical studies for treating myeloproliferative neoplasms (MPNs) and other hematologic cancers. Clinical trials have indicated that IMG-7289 can reduce disease burden and improve symptoms in patients with MPNs.
The benefits of IMG-7289 include its potential to induce remission in refractory or relapsed cancers and its oral administration route, which enhances patient compliance. However, risks include myelosuppression, gastrointestinal issues, and potential off-target effects, necessitating precise dosing and monitoring [70].
Recent developments in LSD1 inhibitors
Research has expanded the use of LSD1 inhibitors to various cancers beyond hematologic malignancies. Combination therapies involving LSD1 inhibitors and other anticancer agents, such as immune checkpoint inhibitors and targeted therapies, have shown enhanced efficacy in preclinical studies.
Clinical trials are ongoing to evaluate these combinations’ safety and effectiveness in solid tumors and other cancer types. These developments highlight the potential of LSD1 inhibitors as part of multi-modal cancer therapy [70].
CRISPR-based epigenetic editing
CRISPR-based epigenetic editing represents a revolutionary approach to precisely modifying the epigenetic landscape without altering the underlying DNA sequence. This technology leverages the CRISPR-dCas9 system to target specific genomic loci and modify DNA methylation and histone modifications [71].
Mechanism of action
CRISPR-dCas9 technology involves a catalytically inactive form of the Cas9 protein (dCas9) fused with various epigenetic modifiers. The dCas9 protein is guided to specific DNA sequences by a single-guide RNA (sgRNA).
Once targeted, dCas9 can be fused with epigenetic enzymes such as DNA methyltransferases (DNMTs), demethylases (TET1), histone acetyltransferases (p300), or histone deacetylases (HDACs) to modulate the epigenetic marks at the targeted site.
This precise targeting allows for the activation or repression of specific genes, enabling the study and potential therapeutic modification of gene expression [71].
Clinical applications
CRISPR-based epigenetic editing has several promising clinical applications, including cancer therapy, neurodegenerative diseases, and metabolic diseases.
Reactivating silenced tumor suppressor genes or repressing oncogenes to inhibit cancer progression. For example, dCas9-TET1 can be used to demethylate and reactivate tumor suppressor genes, while dCas9-KRAB can add repressive histone marks to silence oncogenes [72].
Modifying the epigenetic regulation of genes involved in neuronal function and survival to treat conditions like Alzheimer’s and Parkinson’s diseases, such as by increasing histone acetylation at the promoter regions of neuroprotective genes using dCas9-p300 [72].
Targeting genes involved in insulin signaling and glucose metabolism to improve metabolic health. dCas9-TET1 or dCas9-DNMT3A can be used to modulate methylation at these genes, enhancing their expression and function [72].
Recent developments
Clinical trials and studies have explored the potential of CRISPR-based epigenetic editing in various diseases. For example, a 2024 by Sven Liesenfelder et al., published in bioRxiv in 2024, demonstrated that targeted demethylation using CRISPR-dCas9-TET1 of age-related genes such as PDE4C improved tissue function and extended lifespan in mice.
The research showed significant and stable changes in DNA methylation, which persisted for over three months, highlighting the potential of epigenetic editing in modulating aging [73].
Combination therapies involving CRISPR-based epigenetic editing and other treatments are also being explored. For example, combining CRISPR-dCas9-based demethylation of immune-related genes with immune checkpoint inhibitors has shown promise in enhancing anti-tumor immune responses [74].
Ongoing clinical trials are evaluating this approach in metastatic melanoma patients, showing early signs of enhanced immune response and tumor regression.
In the future, CRISPR-based epigenetic editing may be used in stem cell therapies to enhance regenerative potential by modifying epigenetic profiles. Preclinical studies have shown that CRISPR-edited stem cells exhibit enhanced regenerative capabilities, and clinical trials are planned for late 2024 to test these cells in patients with chronic heart failure [75].
Additionally, precision medicine approaches are being developed to tailor epigenetic therapies to individual patients based on their unique epigenetic profiles. This personalized approach aims to increase the efficacy and reduce the side effects of treatments for various diseases, including cancer, neurological disorders, and autoimmune diseases [76].
LSD1 inhibitors and CRISPR-based epigenetic editing technologies represent cutting-edge advancements in the field of epigenetic therapies. LSD1 inhibitors, such as ORY-1001 and IMG-7289, have shown promise in treating various cancers by reactivating silenced genes involved in differentiation and apoptosis.
CRISPR-based epigenetic editing offers a precise method to modify the epigenetic landscape, with applications in cancer therapy, neurodegenerative diseases, and metabolic disorders.
Ongoing research and clinical trials continue to refine these approaches, aiming to maximize their therapeutic potential while minimizing adverse effects. As our understanding of epigenetic regulation deepens, these innovative therapies will play a crucial role in personalized medicine and the treatment of complex diseases.
The development of epigenetic biomarkers and personalized medicine approaches has significantly advanced our understanding of aging and disease mechanisms. These tools offer precise insights into biological age, disease prognosis, and treatment responses, paving the way for tailored therapeutic strategies.
Epigenetic clocks
Epigenetic clocks are tools that estimate biological age based on DNA methylation patterns. These clocks provide a more accurate measure of aging compared to chronological age by reflecting the cumulative effects of genetic, environmental, and lifestyle factors on the genome.
The Horvath clock, developed by Steve Horvath in 2013, is a first-generation epigenetic clock that utilizes DNA methylation data from various tissues. It has been widely validated across different populations and is considered a robust predictor of biological age [76].
PhenoAge was developed in 2018 and refined in 2023. It incorporates clinical biomarkers along with DNA methylation data to provide a comprehensive assessment of aging. It includes factors such as blood biomarkers, immune function, and physical fitness levels, allowing for predictions about the onset of age-related diseases [77].
GrimAge was developed by Horvath and colleagues, and it predicts lifespan and healthspan by measuring DNA methylation markers associated with smoking, inflammation, and other risk factors. The 2022 version includes additional markers for inflammatory and metabolic processes, further refining its predictive capabilities [78].
DNAmFitAge focuses on physical fitness and muscle function, using methylation patterns associated with exercise and physical activity levels. It is beneficial for designing personalized fitness programs and assessing the impact of exercise interventions on biological age.
Research in 2023-2024 has validated its accuracy in predicting fitness-related aging and recovery from exercise-induced stress [79].
Recent developments and validation studies
Validation studies have demonstrated the accuracy of these new epigenetic clocks across different populations, ages, and health conditions. Large-scale cross-sectional studies have shown high correlations between predicted biological age and clinical health outcomes.
Longitudinal studies tracking individuals over several years have demonstrated that changes in epigenetic age, as measured by these clocks, correlate with changes in health status and mortality risk [80]. These clocks are increasingly being used in clinical settings to assess patient health, monitor disease progression, and evaluate the effectiveness of therapeutic interventions.
In research, they are valuable tools for studying the mechanisms of aging, testing anti-aging treatments, and exploring the impact of environmental and lifestyle factors on biological aging [81].
Technological advances have further improved the accuracy and robustness of these epigenetic clocks. Advanced machine learning techniques have been applied to refine the predictive algorithms, and high-throughput sequencing technologies allow for the comprehensive analysis of DNA methylation patterns, enhancing the precision of epigenetic age predictions.
Predictive tools
Predictive tools using epigenetic biomarkers enable personalized treatment plans by assessing patient responses to therapies and tailoring interventions accordingly. These biomarkers are specific epigenetic signatures associated with disease states, enabling more accurate diagnosis, prognosis, and monitoring of treatment efficacy [82].
Use of biomarkers for personalized treatment plans
Biomarkers identified through epigenetic studies can predict how patients will respond to certain therapies, allowing for personalized treatment strategies.
For instance, DNA methylation biomarkers have been used to stratify patients with myelodysplastic syndromes (MDS) and predict their response to DNMT inhibitors, improving treatment outcomes [83].
Similarly, histone modification patterns and non-coding RNA profiles have been used to tailor treatments for cancers and neurodegenerative diseases [84, 85].
Recent studies and clinical applications
Studies have highlighted the clinical applications of these predictive tools. For example, a 2023 study identified DNA methylation biomarkers that predict response to DNMT inhibitors in MDS patients, improving treatment stratification and outcomes [86].
Another study demonstrated that non-coding RNA profiles could predict patient responses to HDAC inhibitors in cancer therapy, allowing for more targeted and effective treatments.
These advancements underscore the potential of epigenetic biomarkers in developing personalized medicine approaches that optimize therapeutic efficacy while minimizing side effects.
Epigenetic drugs and biomarkers have revolutionized modern medicine by providing new avenues for treating complex diseases and improving patient outcomes. DNMT inhibitors, HDAC inhibitors, and other epigenetic drugs have shown efficacy in reactivating silenced genes and modulating gene expression, offering new hope for patients with cancer, neurodegenerative diseases, and other conditions.
Epigenetic biomarkers, including advanced epigenetic clocks, have enabled more precise assessments of biological age, disease risk, and treatment responses, facilitating personalized medicine approaches [84].
The future of personalized epigenetic therapies holds immense potential. Ongoing research aims to refine these therapies, develop new epigenetic drugs, and enhance predictive tools for better patient stratification and treatment planning.
Advances in CRISPR-based epigenetic editing and other innovative technologies promise to provide even more precise and effective interventions. As our understanding of the epigenetic landscape grows, these therapies will likely play an increasingly vital role in personalized medicine, offering tailored treatments that improve health outcomes and extend healthy lifespan.
The integration of epigenetic insights into clinical practice will transform how we approach disease prevention, diagnosis, and treatment, paving the way for a new era of healthcare.
Conclusion
Epigenetic alterations, including DNA methylation, histone modifications, and non-coding RNA regulation, play a critical role in aging by influencing gene expression without changing the underlying DNA sequence. Advances in epigenetic reprogramming technologies, particularly CRISPR-based epigenetic editing, have shown promising results in reversing age-related changes, treating cancer, and enhancing cognitive function.
Physical activity, stress management, and dietary habits significantly impact the epigenetic landscape. Exercise-induced epigenetic changes, stress reduction techniques, and bioactive compounds from food can modulate gene expression and improve health outcomes.
The gut microbiome influences epigenetic regulation by producing metabolites like short-chain fatty acids, vitamins, and bile acids, which can affect DNA methylation and histone modifications.
New epigenetic clocks, such as Horvath’s second-generation clock and the PhenoAge clock, provide accurate biological age and healthspan predictions, offering valuable tools for personalized medicine and anti-aging interventions.
Recent approvals and ongoing trials of epigenetic drugs, such as DNMT inhibitors, HDAC inhibitors, and BET inhibitors, are transforming cancer treatment and other diseases. These therapies, combined with new diagnostic tools, are enhancing patient care and outcomes.
Future research in epigenetic editing will focus on increasing precision, reducing off-target effects, and expanding applications to more diseases. Researchers aim to refine these technologies to achieve durable and targeted epigenetic modifications. Emphasis will be placed on personalized approaches, using individual epigenetic profiles to tailor treatments.
This involves developing more sophisticated biomarkers and predictive tools to optimize therapeutic strategies. Exploring the synergistic effects of combining epigenetic drugs with other treatments, such as immunotherapies and targeted therapies, will be a key area of focus. These combinations have the potential to enhance efficacy and overcome resistance mechanisms.
Further investigation into the interactions between the gut microbiome and the epigenome will provide deeper insights into how diet, lifestyle, and microbial composition influence aging and disease.
This research could lead to microbiome-based interventions to modulate epigenetic marks. Research into the epigenetic mechanisms underlying neurodegenerative diseases will expand, with a focus on developing therapies that can modify epigenetic marks to improve cognitive function and slow disease progression.
As epigenetic therapies advance, addressing ethical and regulatory challenges will be crucial. Ensuring equitable access, maintaining genetic privacy, and understanding the long-term effects of epigenetic modifications will be important considerations.
Epigenetic therapies have the potential to revolutionize disease prevention and management by targeting the root causes of epigenetic dysregulation. This could lead to more effective treatments for cancer, neurodegenerative diseases, metabolic disorders, and autoimmune conditions. By modulating epigenetic marks, it may be possible to extend healthspan, the period of life spent in good health.
Epigenetic interventions could delay the onset of age-related diseases, improve quality of life, and reduce healthcare costs associated with aging populations. The integration of epigenetic biomarkers into clinical practice will enable personalized healthcare approaches, allowing for more accurate diagnoses, tailored treatments, and better monitoring of disease progression and treatment response.
Hopefully, epigenetic research will contribute to the development of preventive medicine strategies, identifying individuals at risk for certain diseases based on their epigenetic profiles and implementing early interventions to mitigate these risks.
Insights from epigenetic research could inform public health policies, emphasizing the importance of lifestyle factors, such as diet, exercise, and stress management, in maintaining optimal epigenetic health and preventing age-related diseases.
Epigenetic research is at the forefront of understanding the complexities of aging and developing innovative therapies to combat age-related diseases. Advances in epigenetic reprogramming, personalized medicine, and lifestyle interventions hold the promise of extending healthspan and improving overall well-being.
As our knowledge of the epigenome continues to expand, the potential for transformative impacts on healthcare and longevity becomes increasingly evident. By addressing ethical and regulatory challenges and continuing to explore the intricate interplay between genetics, environment, and lifestyle, we can pave the way for a healthier, longer-lived future.
Literature
[1] Willbanks, A.; Leary, M.; Greenshields, M.; Tyminski, C.; Heerboth, S.; Lapinska, K.; Haskins, K.; Sarkar, S. The Evolution of Epigenetics: From Prokaryotes to Humans and Its Biological Consequences. Genet Epigenet 2016, 1, 25–36.
[2] Hackett, J.A.; Azim Surani, M. DNA Methylation Dynamics during the Mammalian Life Cycle. Philosophical Transactions of the Royal Society B: Biological Sciences 2013, 368.
[3] López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194.
[4] López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278.
[5] Fülöp, T.; Larbi, A.; Witkowski, J.M. Human Inflammaging. Gerontology 2019, 65, 495–504.
[6] Lu, Y.; Brommer, B.; Tian, X.; Krishnan, A.; Meer, M.; Wang, C.; Vera, D.L.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reprogramming to Recover Youthful Epigenetic Information and Restore Vision. Nature 2020, 588, 124–129.
[7] Xie, N.; Zhou, Y.; Sun, Q.; Tang, B. Novel Epigenetic Techniques Provided by the CRISPR/Cas9 System. Stem Cells Int 2018, 2018, 7834175.
[8] Yang, J.H.; Petty, C.A.; Dixon-McDougall, T.; Lopez, M.V.; Tyshkovskiy, A.; Maybury-Lewis, S.; Tian, X.; Ibrahim, N.; Chen, Z.; Griffin, P.T.; et al. Chemically Induced Reprogramming to Reverse Cellular Aging. Aging (Albany NY) 2023, 15, 5966.
[9] Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676.
[10] Ocampo, A.; Reddy, P.; Martinez-Redondo, P.; Platero-Luengo, A.; Hatanaka, F.; Hishida, T.; Li, M.; Lam, D.; Kurita, M.; Beyret, E.; et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 2016, 167, 1719-1733.e12.
[11] Park, H.; Shin, J.; Kim, Y.; Saito, T.; Saido, T.C.; Kim, J. CRISPR/DCas9-Dnmt3a-Mediated Targeted DNA Methylation of APP Rescues Brain Pathology in a Mouse Model of Alzheimer’s Disease. Transl Neurodegener 2022, 11, 1–12.
[12] Kornicka, K.; Marycz, K.; Marędziak, M.; Tomaszewski, K.A.; Nicpoń, J. The Effects of the DNA Methyltranfserases Inhibitor 5-Azacitidine on Ageing, Oxidative Stress and DNA Methylation of Adipose Derived Stem Cells. J Cell Mol Med 2017, 21, 387–401.
[13] Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M.; et al. The Achilles’ Heel of Senescent Cells: From Transcriptome to Senolytic Drugs. Aging Cell 2015, 14, 644–658.
[14] Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol 2013, 14, 3156.
[15] Widmann, M.; Nieß, A.M.; Munz, B. Physical Exercise and Epigenetic Modifications in Skeletal Muscle. Sports Medicine 2019, 49, 509–523.
[16] Lochmann, T.L.; Thomas, R.R.; Bennett, J.P.; Taylor, S.M. Epigenetic Modifications of the PGC-1α Promoter during Exercise Induced Expression in Mice. PLoS One 2015, 10, e0129647.
[17] Choi, Y.S.; Kim, S.; Lee, H.K.; Lee, K.-U.; Pak, Y.K. In Vitro Methylation of Nuclear Respiratory Factor-1 Binding Site Suppresses the Promoter Activity of Mitochondrial Transcription Factor A q.
[18] Sillanpää, E.; Ollikainen, M.; Kaprio, J.; Wang, X.; Leskinen, T.; Kujala, U.M.; Törmäkangas, T. Leisure-Time Physical Activity and DNA Methylation Age – A Twin Study. Clin Epigenetics 2019, 11, 1–8.
[19] Cechinel, L.R.; Basso, C.G.; Bertoldi, K.; Schallenberger, B.; de Meireles, L.C.F.; Siqueira, I.R. Treadmill Exercise Induces Age and Protocol-Dependent Epigenetic Changes in Prefrontal Cortex of Wistar Rats. Behavioural Brain Research 2016, 313, 82–87.
[20] Wilker, S.; Vukojevic, V.; Schneider, A.; Pfeiffer, A.; Inerle, S.; Pauly, M.; Elbert, T.; Papassotiropoulos, A.; de Quervain, D.; Kolassa, I.T. Epigenetics of Traumatic Stress: The Association of NR3C1 Methylation and Posttraumatic Stress Disorder Symptom Changes in Response to Narrative Exposure Therapy. Translational Psychiatry 2023 13:1 2023, 13, 1–7.
[21] Watkeys, O.J.; Kremerskothen, K.; Quidé, Y.; Fullerton, J.M.; Green, M.J. Glucocorticoid Receptor Gene (NR3C1) DNA Methylation in Association with Trauma, Psychopathology, Transcript Expression, or Genotypic Variation: A Systematic Review. Neurosci Biobehav Rev 2018, 95, 85–122.
[22] Venditti, S.; Verdone, L.; Reale, A.; Vetriani, V.; Caserta, M.; Zampieri, M. Molecules of Silence: Effects of Meditation on Gene Expression and Epigenetics. Front Psychol 2020, 11, 544346.
[23] Verdone, L.; Caserta, M.; Ben-Soussan, T.D.; Venditti, S. On the Road to Resilience: Epigenetic Effects of Meditation. Vitam Horm 2023, 122, 339–376, doi:10.1016/BS.VH.2022.12.009.
[24] Kripalani, S.; Pradhan, B.; Gilrain, K.L. The Potential Positive Epigenetic Effects of Various Mind-Body Therapies (MBTs): A Narrative Review. J Complement Integr Med 2022, 19, 827–832.
[25] Zimmer, P.; Schenk, A.; Bloch, W. Epigenetics in Exercise Science and Sports Medicine. Med Epigenet 2016, 515–530, doi:10.1016/B978-0-12-803239-8.00029-6.
[26] Franzago, M.; Pilenzi, L.; Di Rado, S.; Vitacolonna, E.; Stuppia, L. The Epigenetic Aging, Obesity, and Lifestyle. Front Cell Dev Biol 2022, 10, 985274.
[27] Choi, S.W.; Claycombe, K.J.; Alfredo Martinez, J.; Friso, S.; Schalinske, K.L. Nutritional Epigenomics: A Portal to Disease Prevention. Advances in Nutrition 2013, 4, 530–532.
[28] Auclair, G.; Weber, M. Mechanisms of DNA Methylation and Demethylation in Mammals. Biochimie 2012, 94, 2202–2211.
[29] Maugeri, A.; Barchitta, M. How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies. Medicina (B Aires) 2020, 56, 1–26.
[30] Ayissi, V.B.O.; Ebrahimi, A.; Schluesenner, H. Epigenetic Effects of Natural Polyphenols: A Focus on SIRT1-Mediated Mechanisms. Mol Nutr Food Res 2014, 58, 22–32.
[31] Yousef, M.; Vlachogiannis, I.A.; Tsiani, E. Effects of Resveratrol against Lung Cancer: In Vitro and in Vivo Studies. Nutrients 2017, 9, 1–14, doi:10.3390/nu9111231.
[32] Özyalçin, B.; Sanlier, N. The Effect of Diet Components on Cancer with Epigenetic Mechanisms. Trends Food Sci Technol 2020, 102, 138–145.
[33] Teiten, M.H.; Dicato, M.; Diederich, M. Curcumin as a Regulator of Epigenetic Events. Mol Nutr Food Res 2013, 57, 1619–1629.
[34] He, L.; Han, M.; Farrar, S.; Ma, X. Editorial: Impacts and Regulation of Dietary Nutrients on Gut Microbiome and Immunity. Protein Pept Lett 2017, 24, 380–381.
[35] Watson, M.M.; van der Giezen, M.; Søreide, K. Gut Microbiome Influence on Human Epigenetics, Health, and Disease. Handbook of Epigenetics: The New Molecular and Medical Genetics, Third Edition 2023, 669–686.
[36] van de Wouw, M.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain Axis: Modulator of Host Metabolism and Appetite. J Nutr 2017, 147, 727–745.
[37] Stilling, R.M.; van de Wouw, M.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. The Neuropharmacology of Butyrate: The Bread and Butter of the Microbiota-Gut-Brain Axis? Neurochem Int 2016, 99, 110–132.
[38] Chen, X.; D’Souza, R.; Hong, S.-T. The Role of Gut Microbiota in the Gut-Brain Axis: Current Challenges and Perspectives. Protein Cell 2013, 4, 403–414.
[39] Majchrzak-Celinska, A.; Warych, A.; Szoszkiewicz, M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes 2021, Vol. 12, Page 208 2021, 12, 208.
[40] Heerboth, S.; Lapinska, K.; Snyder, N.; Leary, M.; Rollinson, S.; Sarkar, S. Use of Epigenetic Drugs in Disease: An Overview. Genet Epigenet 2014, 1, 9–19.
[41] Mehdipour, P.; Chen, R.; De Carvalho, D.D. The next Generation of DNMT Inhibitors. Nature Cancer 2021 2:10 2021, 2, 1000–1001.
[42] Kaminskas, E.; Farrell, A.T.; Wang, Y.-C.; Sridhara, R.; Pazdur, R. FDA Drug Approval Summary: Azacitidine (5-Azacytidine, VidazaTM) for Injectable Suspension. Oncologist 2005, 10, 176–182.
[43] Rubio-San-Simón, A.; van Eijkelenburg, N.K.A.; Hoogendijk, R.; Hasle, H.; Niemeyer, C.M.; Dworzak, M.N.; Zecca, M.; Lopez-Yurda, M.; Janssen, J.M.; Huitema, A.D.R.; et al. Azacitidine (Vidaza®) in Pediatric Patients with Relapsed Advanced MDS and JMML: Results of a Phase I/II Study by the ITCC Consortium and the EWOG-MDS Group (Study ITCC-015). Pediatric Drugs 2023, 25, 719–728.
[44] Nieto, M.; Demolis, P.; Béhanzin, E.; Moreau, A.; Hudson, I.; Flores, B.; Stemplewski, H.; Salmonson, T.; Gisselbrecht, C.; Bowen, D.; et al. The European Medicines Agency Review of Decitabine (Dacogen) for the Treatment of Adult Patients With Acute Myeloid Leukemia: Summary of the Scientific Assessment of the Committee for Medicinal Products for Human Use. Oncologist 2016, 21, 692–700.
[45] Luo, N.; Sugiura, A.; Balko, J.M. Therapeutic Potential of DNA Methyltransferase Inhibitors with Immune Checkpoint Inhibitor Therapy in Breast Cancer. Cell Stress 2018, 2, 69.
[46] Kim, D.Y.; Shin, D.Y.; Oh, S.; Kim, I.; Kim, E.J. Gene Expression and DNA Methylation Profiling Suggest Potential Biomarkers for Azacitidine Resistance in Myelodysplastic Syndrome. Int J Mol Sci 2024, 25.
[47] Majchrzak-Celinska, A.; Warych, A.; Szoszkiewicz, M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes 2021, Vol. 12, Page 208 2021, 12, 208.
[48] Pereira, M.; Cruz, M.T.; Fortuna, A.; Bicker, J. Restoring the Epigenome in Alzheimer’s Disease: Advancing HDAC Inhibitors as Therapeutic Agents. Drug Discov Today 2024, 29, 104052.
[49] Wawruszak, A.; Borkiewicz, L.; Okon, E.; Kukula-Koch, W.; Afshan, S.; Halasa, M. Vorinostat (SAHA) and Breast Cancer: An Overview. Cancers 2021, Vol. 13, Page 4700 2021, 13, 4700.
[50] Iyer, S.P.; Huen, A.; Ai, W.Z.; Jagadeesh, D.; Lechowicz, M.J.; Okada, C.; Feldman, T.A.; Ghione, P.; Alderuccio, J.P.; Champion, R.; et al. Safety and Efficacy of Tenalisib in Combination with Romidepsin in Patients with Relapsed/Refractory T-Cell Lymphoma: Results from a Phase I/II Open-Label Multicenter Study. Haematologica 2024, 109, 209.
[51] Dunn, A.; Takebe, N.; Chen, A.; Kummar, S.; Piekarz, R.; Kiesel, B.; Moore, N.; Doroshow, J.; Beumer, J.H.; Gobburu, J.V.S. The Effect of Liver Dysfunction on the Pharmacokinetic Disposition of Belinostat and Its Five Metabolites in Patients with Advanced Cancers. Cancer Chemother Pharmacol 2024, 1–11.
[52] Mackay, H.J.; Hirte, H.; Colgan, T.; Covens, A.; MacAlpine, K.; Grenci, P.; Wang, L.; Mason, J.; Pham, P.A.; Tsao, M.S.; et al. Phase II Trial of the Histone Deacetylase Inhibitor Belinostat in Women with Platinum Resistant Epithelial Ovarian Cancer and Micropapillary (LMP) Ovarian Tumours. Eur J Cancer 2010, 46, 1573–1579.
[53] Yao, D.; Li, C.; Jiang, J.; Huang, J.; Wang, J.; He, Z.; Zhang, J. Design, Synthesis and Biological Evaluation of Novel HDAC Inhibitors with Improved Pharmacokinetic Profile in Breast Cancer. Eur J Med Chem 2020, 205, 112648, doi:10.1016/J.EJMECH.2020.112648.
[54] Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 Is a Microtubule-Associated Deacetylase. Nature 2002 417:6887 2002, 417, 455–458.
[55] Médard, G.; Sheltzer, J.M. Ricolinostat Is Not a Highly Selective HDAC6 Inhibitor. Nature Cancer 2023 4:6 2023, 4, 807–808.
[56] Vogl, D.T.; Raje, N.S.; Jagannath, S.; Richardson, P.G.; Hari, P.; Orlowski, R.Z.; Supko, J.; Tamang, D.; Jones, S.S.; Wheeler, C.; et al. Ricolinostat (ACY-1215), the First Selective HDAC6 Inhibitor, in Combination with Bortezomib and Dexamethasone in Patients with Relapsed or Relapsed-and-Refractory Multiple Myeloma: Phase 1b Results (ACY-100 Study). Blood 2015, 126, 1827.
[57] Utsunomiya, A.; Izutsu, K.; Jo, T.; Yoshida, S.; Tsukasaki, K.; Ando, K.; Choi, I.; Imaizumi, Y.; Kato, K.; Kurosawa, M.; et al. Oral Histone Deacetylase Inhibitor Tucidinostat (HBI-8000) in Patients with Relapsed or Refractory Adult T-Cell Leukemia/Lymphoma: Phase IIb Results. Cancer Sci 2022, 113, 2778–2787.
[58] Yang, H.M.; Lee, C.; Min, J.; Ha, N.; Bae, D.; Nam, G.; Park, H.J. Development of a Tetrahydroindazolone-Based HDAC6 Inhibitor with in-Vivo Anti-Arthritic Activity. Bioorg Med Chem 2024, 99, 117587.
[59] Simões-Pires, C.; Zwick, V.; Nurisso, A.; Schenker, E.; Carrupt, P.A.; Cuendet, M. HDAC6 as a Target for Neurodegenerative Diseases: What Makes It Different from the Other HDACs? Mol Neurodegener 2013, 8, 1–16.
[60] Marzochi, L.L.; Cuzziol, C.I.; Nascimento Filho, C.H.V. Do; dos Santos, J.A.; Castanhole-Nunes, M.M.U.; Pavarino, É.C.; Guerra, E.N.S.; Goloni-Bertollo, E.M. Use of Histone Methyltransferase Inhibitors in Cancer Treatment: A Systematic Review. Eur J Pharmacol 2023, 944, 175590.
[61] Duan, R.; Du, W.; Guo, W. EZH2: A Novel Target for Cancer Treatment. J Hematol Oncol 2020, 13.
[62] Kang, N.; Eccleston, M.; Clermont, P.L.; Latarani, M.; Male, D.K.; Wang, Y.; Crea, F. EZH2 Inhibition: A Promising Strategy to Prevent Cancer Immune Editing. Epigenomics 2020, 12, 1457–1476.
[63] Straining, PharmD, R.; Eighmy, PharmD, W. Tazemetostat: EZH2 Inhibitor. J Adv Pract Oncol 2022, 13, 158.
[64] Kazansky, Y.; Cameron, D.; Mueller, H.S.; Demarest, P.; Zaffaroni, N.; Arrighetti, N.; Zuco, V.; Kuwahara, Y.; Somwar, R.; Ladanyi, M.; et al. Overcoming Clinical Resistance to EZH2 Inhibition Using Rational Epigenetic Combination Therapy. Cancer Discov 2024, 14, 965–981.
[65] Paolini, R.L.; Souroullas, G.P. The Cell Cycle: A Key to Unlock EZH2-Targeted Therapy Resistance. Cancer Discov 2024, 14, 903–905.
[66] Noguchi-Yachide, T. BET Bromodomain as a Target of Epigenetic Therapy. Chem Pharm Bull (Tokyo). 2016, 64, 540–547.
[67] Meng, S.; Zhang, L.; Tang, Y.; Tu, Q.; Zheng, L.; Yu, L.; Murray, D.; Cheng, J.; Kim, S.H.; Zhou, X.; et al. BET Inhibitor JQ1 Blocks Inflammation and Bone Destruction. Journal of Dental Research 2014, 93, 657–662.
[68] Divakaran, A.; Harki, D.A.; Pomerantz, W.C.K. Recent Progress and Structural Analyses of Domain-Selective BET Inhibitors. Med Res Rev 2023, 43, 972–1018.
[69] Baby, S.; Shinde, S.D.; Kulkarni, N.; Sahu, B. Lysine-Specific Demethylase 1 (LSD1) Inhibitors: Peptides as an Emerging Class of Therapeutics. ACS Chem Biol 2023, 18, 2144–2155.
[70] Liu, H.M.; Zhou, Y.; Chen, H.X.; Wu, J.W.; Ji, S.K.; Shen, L.; Wang, S.P.; Liu, H.M.; Liu, Y.; Dai, X.J.; et al. LSD1 in Drug Discovery: From Biological Function to Clinical Application. Med Res Rev 2024, 44, 833–866.
[71] Kolanu, N.D. CRISPR–Cas9 Gene Editing: Curing Genetic Diseases by Inherited Epigenetic Modifications. Glob Med Genet 2024, 11, 113–122.
[72] Fadul, S.M.; Arshad, A.; Mehmood, R. CRISPR-Based Epigenome Editing: Mechanisms and Applications. Epigenomics 2023, 15, 1137–1155.
[73] Liesenfelder, S.; Mabrouk, M.H.E.; Iliescu, J.; Baranda, M.V.; Mizi, A.; Wessiepe, M.; Papantonis, A.; Wagner, W. Epigenetic Editing at Individual Age-Associated CpGs Affects the Genome-Wide Epigenetic Aging Landscape. bioRxiv 2024, 2024.06.04.597161.
[74] Baretti, M.; Yarchoan, M. Epigenetic Modifiers Synergize with Immune-Checkpoint Blockade to Enhance Long-Lasting Antitumor Efficacy. J Clin Invest 2021, 131.
[75] Pulecio, J.; Verma, N.; Mejía-Ramírez, E.; Huangfu, D.; Raya, A. CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell 2017, 21, 431–447.
[76] Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol 2013, 14, 1–20..
[77] Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An Epigenetic Biomarker of Aging for Lifespan and Healthspan. Aging (Albany NY) 2018, 10, 573.
[78] Lu, A.T.; Binder, A.M.; Zhang, J.; Yan, Q.; Reiner, A.P.; Cox, S.R.; Corley, J.; Harris, S.E.; Kuo, P.L.; Moore, A.Z.; et al. DNA Methylation GrimAge Version 2. Aging (Albany NY) 2022, 14, 9484.
[79] McGreevy, K.M.; Radak, Z.; Torma, F.; Jokai, M.; Lu, A.T.; Belsky, D.W.; Binder, A.; Marioni, R.E.; Ferrucci, L.; Pośpiech, E.; et al. DNAmFitAge: Biological Age Indicator Incorporating Physical Fitness. Aging (Albany NY) 2023, 15, 3904.
[80] Vetter, V.M.; Kalies, C.H.; Sommerer, Y.; Spira, D.; Drewelies, J.; Regitz-Zagrosek, V.; Bertram, L.; Gerstorf, D.; Demuth, I. Relationship Between 5 Epigenetic Clocks, Telomere Length, and Functional Capacity Assessed in Older Adults: Cross-Sectional and Longitudinal Analyses. The Journals of Gerontology: Series A 2022, 77, 1724–1733.
[81] Moqri, M.; Herzog, C.; Poganik, J.R.; Ying, K.; Justice, J.N.; Belsky, D.W.; Higgins-Chen, A.T.; Chen, B.H.; Cohen, A.A.; Fuellen, G.; et al. Validation of Biomarkers of Aging. Nature Medicine 2024 30:2 2024, 30, 360–372.
[82] Bornhorst, J. An Introduction to Personalized Medicine. Ther Drug Monit 2024, 331–354.
[83] Reilly, B.; Tanaka, T.N.; Diep, D.; Yeerna, H.; Tamayo, P.; Zhang, K.; Bejar, R. DNA Methylation Identifies Genetically and Prognostically Distinct Subtypes of Myelodysplastic Syndromes. Blood Adv 2019, 3, 2845–2858.
[84] Parikh, D.; Shah, M. A Comprehensive Study on Epigenetic Biomarkers in Early Detection and Prognosis of Alzheimer’s Disease. Biomedical Analysis 2024, 1, 138–153.
[85] Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Non-Coding RNA 2021, Vol. 7, Page 47 2021, 7, 47.
[86] Kagan, A.B.; Garrison, D.A.; Anders, N.M.; Webster, J.A.; Baker, S.D.; Yegnasubramanian, S.; Rudek, M.A. DNA Methyltransferase Inhibitor Exposure–Response: Challenges and Opportunities. Clin Transl Sci 2023, 16, 1309,




