Creating a Noise Clock to Measure Biological Age
- Current epigenetic clocks might not be measuring the right things.

Publishing in Aging, the Conboy research lab has outlined the problems with existing machine learning-based clocks and created a new clock based on epigenetic noise.
The problem with linear regression
This paper begins with a description of the elastic net, an artificial intelligence algorithm that powers the majority of biological clocks, including the Horvath biological clock [1], the Pace of Aging clock [2], and the mortality predictor GrimAge [3]. This algorithm is programmed to find the most likely linear result [4].
However, these researchers note that actual epigenetic aging is not necessarily linear and that these clocks do not, and cannot, understand the underlying biology. Instead, the current epigenetic clocks are based on the methylation of the cytosine nucleotide on specific parts of the genome, as measured by optical probes. Many of these locations are isolated and scattered throughout the genome.
Specific age-related biomarkers, such as senescent cells and inflammation, are impossible to directly measure using this method, and the researchers also note that elastic net outputs are not normalized against one another. Subtle differences between the ways that experiments are conducted mean that the output of an elastic net measurement is not necessarily replicable between experiments [5]. Similarly, data-skewing artifacts can be introduced both by technical problems and genomic instability in older people [6].
Therefore, these researchers took a closer examination of these clocks in order to determine their true value as predictors of biological aging.
A search for true accuracy
The first experiment was a straightforward evaluation of clock accuracy. While there were strong correlations at the 18-30 age range, which the authors ascribe to immune system maturity, and at the 80-100 age range at which most people rapidly deteriorate, the accuracy of methylation-based clocks is considerably reduced at other age ranges. The authors note that this is likely due to non-linear aging effects, with healthy plateaus and periods of sharp decline due to the disrepair of multiple tissues [7].
The researchers then evaluated the clocks’ relationship to various diseases. They found that there was significant overlap between diseased groups and healthy groups in the elastic net results, even in diseases such as inflammaging-increasing arthritis and Werner syndrome, along with Down syndrome, whose sufferers have pathologically juvenile blood phenotypes. Another clock showed Parkinson’s patients to be far younger than their healthy counterparts.
Chronology or biology?
In the Conboys’ view, such clocks are predicting chronology more than biology. To illustrate their point, they created a methylation clock based on the US population over time: a proxy for pure chronology. A similar clock also used 149 cytosines to accurately predict chronological age.
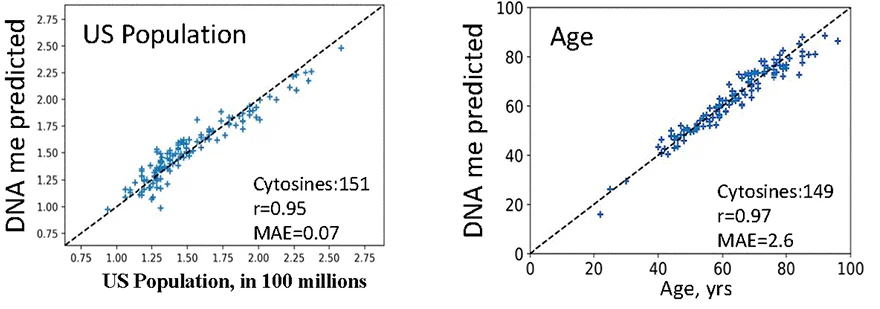
The researchers found this to be more of a problem with the elastic net than the raw array data. Using a Uniform Manifold Approximation and Projection method, they found that they could accurately use the full array of measured cytosines to differentiate healthy people and sick people, while a clock based on the elastic net does not. This, the researchers contend, is due to the elastic net disregarding health and biological factors in favor of the chronological result it was told to create.
A more in-depth analysis found that elastic net-based clocks do not choose cytosines based on their actual change with biological age. When repeatedly removing unchosen cytosines from the clock’s training data, the researchers found that the chosen cytosines would also change.
Such behavior does not agree with a hypothesis that biomarkers should not change when non-biomarkers are deleted from a dataset and supports the null hypothesis that picked cytosines are not biomarkers.
Biological noise as a biomarker
With these problem in mind, the researchers decided to use 1,806 samples to make a better clock. They selected cytosines that, instead of reliably changing with age, normally don’t change at all. Many of the relevant genes were found to serve vital functions in the human body, encompassing such functions as cellular motility and cholesterol regulation. Any changes to these genes, therefore, is the result of biological noise.
The researchers found that their “noise barometer” was correlated strongly with disease. For example, unlike in linear clocks, arthritis patients were found to have significantly higher biological ages compared to their chronological ages. Furthermore, regardless of the tested cytosine, one fact was consistently found to be true: the genome becomes more noisy with age.
This work shifts the dominant paradigm of what is biological age, uncovers that it is not an evenly “ticking clock” or a straight line, and establishes how biological age and risk of disease can be accurately measured. The outcomes minimize the danger to people who might be mistakenly told that they are older or younger and encouraged to take drugs or supplements, based on potential random variation or a normal healthy range, after an assay on a droplet of their blood.
Literature
[1] Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome biology, 14(10), 1-20.
[2] Belsky, D. W., Caspi, A., Corcoran, D. L., Sugden, K., Poulton, R., Arseneault, L., … & Moffitt, T. E. (2022). DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife, 11, e73420.
[3] Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., … & Horvath, S. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (albany NY), 11(2), 303.
[4] Zou, H., & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society Series B: Statistical Methodology, 67(2), 301-320.
[5] Bell, C. G., Lowe, R., Adams, P. D., Baccarelli, A. A., Beck, S., Bell, J. T., … & Rakyan, V. K. (2019). DNA methylation aging clocks: challenges and recommendations. Genome biology, 20, 1-24.
[6] Hop, P. J., Zwamborn, R. A., Hannon, E. J., Dekker, A. M., van Eijk, K. R., Walker, E. M., … & Veldink, J. H. (2020). Cross-reactive probes on Illumina DNA methylation arrays: a large study on ALS shows that a cautionary approach is warranted in interpreting epigenome-wide association studies. NAR Genomics and Bioinformatics, 2(4), lqaa105.
[7] Lai, R. W., Lu, R., Danthi, P. S., Bravo, J. I., Goumba, A., Sampathkumar, N. K., & Benayoun, B. A. (2019). Multi-level remodeling of transcriptional landscapes in aging and longevity. BMB reports, 52(1), 86.







