Intermittent Fasting Induces Changes in Multiple Biomarkers
- The study was on young males practicing fasting during Ramadan.

A study published in Human Nutrition & Metabolism found that prolonged intermittent fasting causes the expression of genes involved in autophagy, the inflammasome, and senescence to change [1].
Fasting your way to better health and longevity?
Previous research has linked fasting to delaying the onset of age-related diseases and longevity along with positive outcomes in several diseases [2]. It has been documented to benefit patients with type 1 and 2 diabetes, cancer, cardiovascular disease, and major depressive disorder [2, 3, 4].
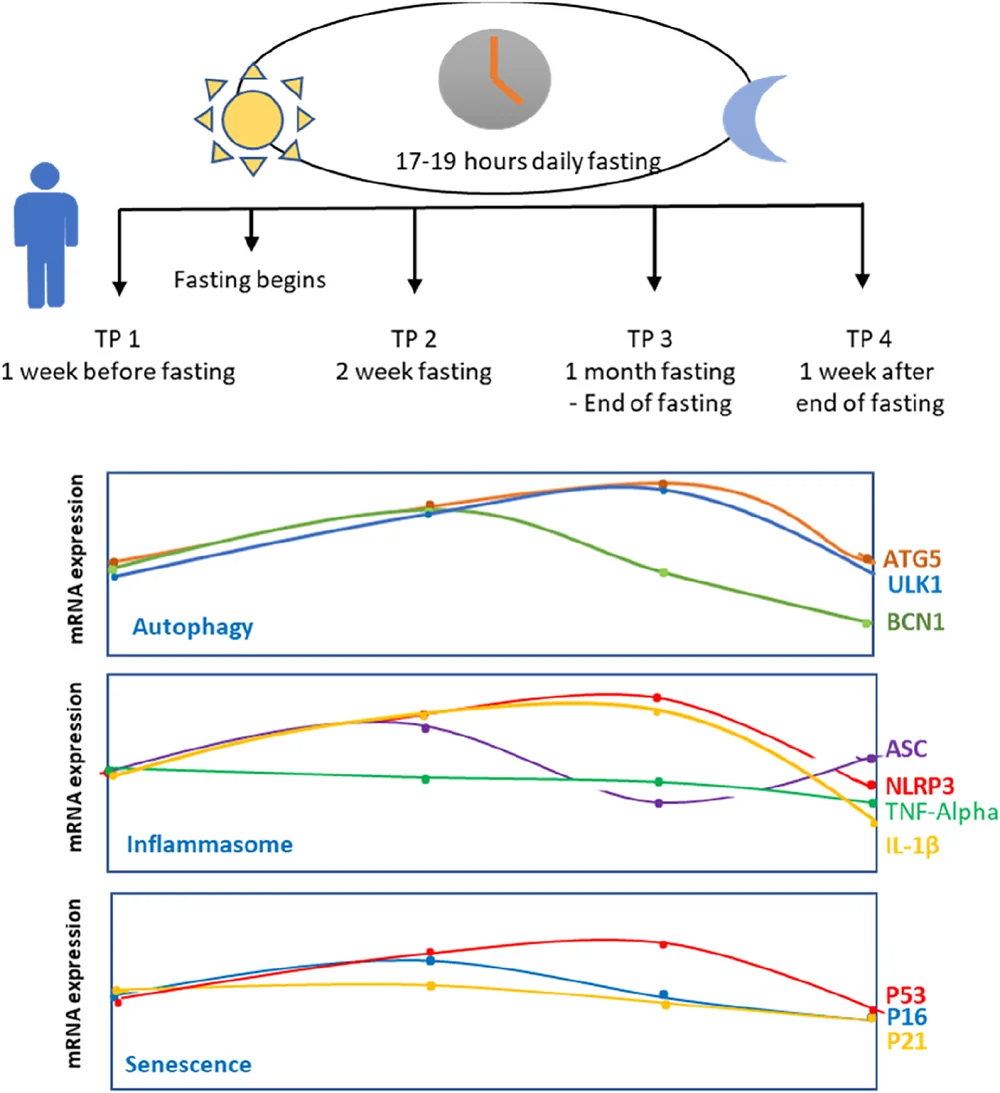
The authors of this paper were particularly interested in what fasting does to the human body on the molecular level, attempting to determine the impacts of prolonged intermittent fasting on health and longevity markers in humans. Therefore, they recruited 25 healthy young men who intended “to fast for the whole month of Ramadan from dawn to dusk.” They measured gene expression levels one week before Ramadan, in the middle of Ramadan, in the last days of Ramadan, and one week after Ramadan.
Fasting induces autophagy
Intermittent fasting activates autophagy, a cellular process through which cells break down their components. Studies have linked the activation of autophagy to longevity, and there are many proteins involved in this process. These researchers tested ULK1, a sensor of nutrient levels and autophagy signals [5]; ATG5, a gene encoding a protein that serves in autophagy induction [6]; and BECN1, a gene encoding a protein necessary in the early steps of autophagy for autophagosome formation [7].
The researchers observed an increase in ULK1 levels caused by fasting two weeks and one month after starting fasting. However, cessation of fasting caused ULK1 to return to its basal levels. Another autophagy protein, ATG5, has shown a similar pattern. The observed pattern is consistent with the function of ULK1 and ATG5 in nutrient sensing and autophagy induction.
BECN1 has shown a different pattern, which included an increase in BECN1 two weeks after starting the fast and a subsequent reduction in its expression levels. Since BECN1’s role in autophagy is more dynamic, this might influence more complex changes in its levels. Additionally, BECN1’s role in apoptosis suggests a hypothesis that “reduction of BECN1 expression level at later time points is to avoid unnecessary apoptosis in healthy individuals” while simultaneously keeping autophagy processes induced, as suggested by the expression of other autophagy genes.
Inflammation and senescence
As part of the immune system, the inflammasome, in response to stimuli, “regulates the activation of many pathways resulting in the secretion of cytokines” [8]. However, the inflammasome can also contribute to inflammaging, a process associated with aging and age-related diseases. The authors measured the expression of genes connected to the inflammasome: NLRP3, a core protein of the inflammasome complex [9]; ASC, a marker of an activated inflammasome complex [10]; and IL-1β, a proinflammatory cytokine [11].
The researchers didn’t find TNF-α levels to change in a statistically significant way, which is contrary to a previous study that found TNF-α upon intermittent fasting.
Other examined genes showed changes in expression. NLRP3 and IL-1β expression was increased two weeks and one month after the start of the intermittent fasting, and the levels decreased one week after the end of fasting. The authors point out that those results contradict other studies. However, they point out that autophagy promotes inflammation in a way that depends on Atg5 [12], connecting it to their results regarding autophagy genes.
On the other hand, they observed that expression of ASC was lower than basal levels one month after the start of intermittent fasting, suggesting that despite higher levels of NLRP3 and IL-1β, most likely caused by ATG5 induction, the inflammasome is not activated. They suggest that prolonged fasting might have activated some non-canonical pathways.
The authors also tested markers of senescence, a cellular process that is a hallmark of aging. Previous research suggests that fasting might reduce senescence by activating autophagy [13]. The senescence markers they used included the senescence mediator p16INK4a, which is essential in senescence initiation through p21 induction [14]. p21 can induce cell cycle arrest, and p53 activation can lead to senescence [15].
The researchers didn’t observe statistically significant changes in the p16INK4a expression levels until the end of their observation. Nevertheless, they observed that p16INK4A expression tends to increase after the start of fasting and then decrease.
The authors observed p21 levels decreasing during and after the fasting. However, those observations are not statistically significant and contradict what was previously reported in animal models. The authors also point out p21’s role as an injury marker required for proliferation and regeneration [16], leading to the hypothesis that p21 levels might be increased during acute fasting but reduced during longer fasting periods.
The final marker was p53 expression, which was increased during fasting. Its levels decreased after fasting cessation. These results align with previous research showing p53 responding to nutrient depletion [17]. p53 can also act as an autophagy activator, which aligns with ATG5 and ULK1 expression during fasting. The authors explain that since they saw an increase in p53 expression but not a significant increase in p16INK4a and p21, they hypothesized this increase to be related to p53’s role in DNA repair but not senescence.
More population variables are needed for future studies
This study has demonstrated that markers of autophagy, inflammasome, and senescence are related in complex ways. While the authors provided many hypothesized explanations, further research is necessary to confirm or refute them.
The authors point out some of this study’s limitations. One is that food intake, physical activity, and sleeping patterns were not recorded, and the authors believe that these variables could have an impact on gene expression patterns. Additionally, only young males were included in this study, making these results questionable for other demographic groups. While the authors recorded the levels of gene expression, the levels of actual proteins may differ, and future studies are needed to assess them.
Literature
[1] Erlangga, Z., Ghashang, S. K., Hamdan, I., Melk, A., Gutenbrunner, C., & Nugraha, B. (2023). The effect of prolonged intermittent fasting on autophagy, inflammasome and senescence genes expressions: An exploratory study in healthy young males. Human Nutrition & Metabolism, 32, 200189.
[2] Longo, V. D., Di Tano, M., Mattson, M. P., & Guidi, N. (2021). Intermittent and periodic fasting, longevity and disease. Nature aging, 1(1), 47–59.
[3] Grajower, M. M., & Horne, B. D. (2019). Clinical Management of Intermittent Fasting in Patients with Diabetes Mellitus. Nutrients, 11(4), 873.
[4] Berthelot, E., Etchecopar-Etchart, D., Thellier, D., Lancon, C., Boyer, L., & Fond, G. (2021). Fasting Interventions for Stress, Anxiety and Depressive Symptoms: A Systematic Review and Meta-Analysis. Nutrients, 13(11), 3947.
[5] Ganley, I. G., Lam, duH., Wang, J., Ding, X., Chen, S., & Jiang, X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. The Journal of biological chemistry, 284(18), 12297–12305.
[6] Zheng, W., Xie, W., Yin, D., Luo, R., Liu, M., & Guo, F. (2019). ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell communication and signaling : CCS, 17(1), 42.
[7] Menon, M. B., & Dhamija, S. (2018). Beclin 1 Phosphorylation – at the Center of Autophagy Regulation. Frontiers in cell and developmental biology, 6, 137.
[8] Zheng, D., Liwinski, T., & Elinav, E. (2020). Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell discovery, 6, 36.
[9] Swanson, K. V., Deng, M., & Ting, J. P. (2019). The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nature reviews. Immunology, 19(8), 477–489.
[10] Schroder, K., & Tschopp, J. (2010). The inflammasomes. Cell, 140(6), 821–832.
[11] Kaneko, N., Kurata, M., Yamamoto, T., Morikawa, S., & Masumoto, J. (2019). The role of interleukin-1 in general pathology. Inflammation and regeneration, 39, 12.
[12] Dupont, N., Jiang, S., Pilli, M., Ornatowski, W., Bhattacharya, D., & Deretic, V. (2011). Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. The EMBO journal, 30(23), 4701–4711.
[13] Shetty, A. K., Kodali, M., Upadhya, R., & Madhu, L. N. (2018). Emerging Anti-Aging Strategies – Scientific Basis and Efficacy. Aging and disease, 9(6), 1165–1184.
[14] Song, S., Lam, E. W., Tchkonia, T., Kirkland, J. L., & Sun, Y. (2020). Senescent Cells: Emerging Targets for Human Aging and Age-Related Diseases. Trends in biochemical sciences, 45(7), 578–592.
[15] Mijit, M., Caracciolo, V., Melillo, A., Amicarelli, F., & Giordano, A. (2020). Role of p53 in the Regulation of Cellular Senescence. Biomolecules, 10(3), 420.
[16] Sturmlechner, I., Zhang, C., Sine, C. C., van Deursen, E. J., Jeganathan, K. B., Hamada, N., Grasic, J., Friedman, D., Stutchman, J. T., Can, I., Hamada, M., Lim, D. Y., Lee, J. H., Ordog, T., Laberge, R. M., Shapiro, V., Baker, D. J., Li, H., & van Deursen, J. M. (2021). p21 produces a bioactive secretome that places stressed cells under immunosurveillance. Science (New York, N.Y.), 374(6567), eabb3420.
[17] Schupp, M., Chen, F., Briggs, E. R., Rao, S., Pelzmann, H. J., Pessentheiner, A. R., Bogner-Strauss, J. G., Lazar, M. A., Baldwin, D., & Prokesch, A. (2013). Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC genomics, 14, 758.








