New System-Specific Epigenetic Clocks Revealed
- Biological systems don't always age at the same rate.

A group of researchers led by Morgan Levine of Altos Labs has created a set of methylation clocks that can detect various aging patterns [1].
The less-than-perfect revolution
The discovery that genome methylation patterns correlate with various processes of aging led to the development of methylation (epigenetic) clocks. They first appeared more than a decade ago [2] and revolutionized geroscience by enabling researchers to gauge the biological age of an organism as opposed to its chronological age. Methylation clocks are now widely used as a proxy to determine the effects of various interventions on lifespan instead of having to wait for organisms to die.
However, the existing clocks are far from perfect. The mechanistic underpinnings of the correlation are often unclear. A clock might give different results on a particular organism than it gives the next day, and this test-retest variability is possibly driven by short-term fluctuations in methylation. Furthermore, while many processes of aging are interlinked, distinct aging patterns exist, as evidenced by the fact that different people acquire different sets of age-related diseases.
Capturing aging patterns
Former Yale professor Morgan Levine, who is now with Altos Labs, is one of the foremost authorities on methylation clocks, and we have interviewed her on the subject. Her group invented the advanced PhenoAge clock [3] along with a method of reducing the variability of existing clocks via principal component analysis. In this new preprint paper, Morgan and her co-authors describe Systems Age, a clock that has been in the works for quite some time. This clock is aimed at discerning between different aging patterns.
The researchers describe several levels of heterogeneity in aging. The first and obvious one is the difference in whole-body aging between individuals: people who have the same chronological age differ in their biological age as measured by today’s methylation clocks. However, there are also more fundamental levels. Subcellular elements, cells, tissues, organs, and systems age differently, which creates distinctive aging subtypes. Systems Age works on these lower levels, capturing interpersonal differences in higher detail.
“Two individuals can have different DNA methylation profiles that produce the exact same epigenetic age as calculated by blood-based epigenetic clocks,” the researchers explain, “yet they may be physiologically deteriorating in entirely different systems.” Accounting for those differences can be immensely helpful for aging research as well as for personalized prevention, detection, and treatment.
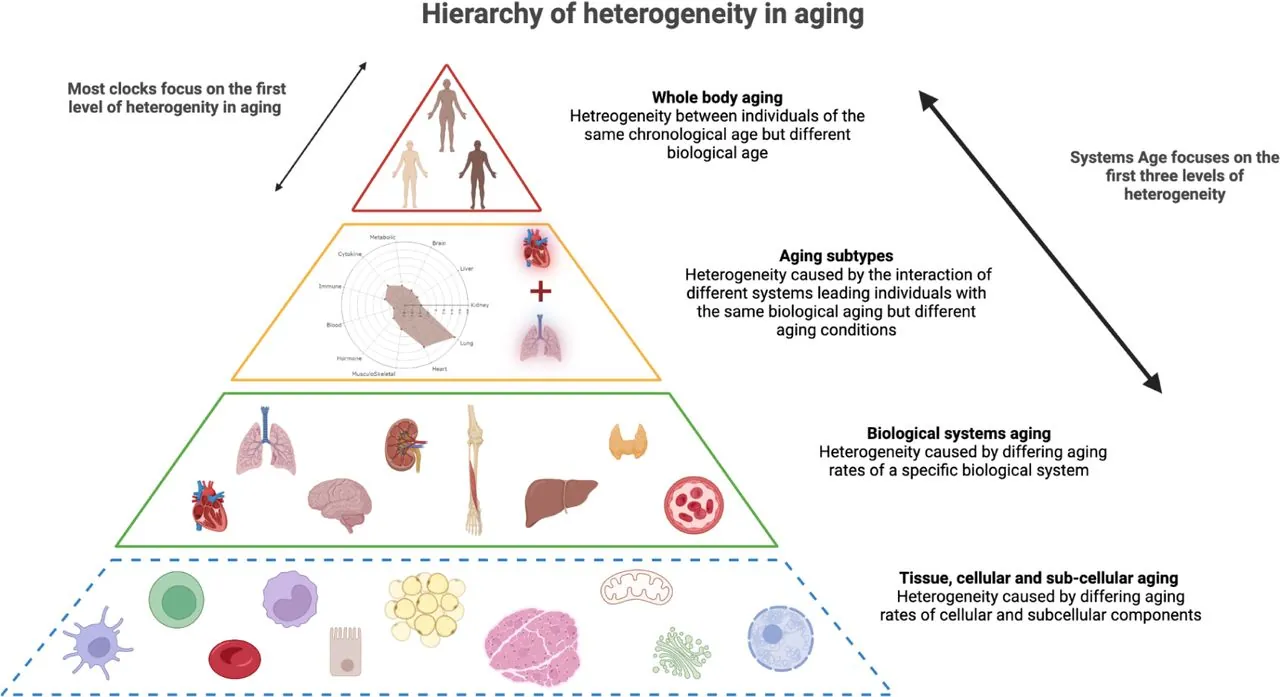
Systems Age was trained on system-specific clinical chemistry and hematology biomarkers from the Health and Retirement study. A mortality prediction model was incorporated as well. The clock includes 11 system-specific scores, such as musculoskeletal, heart, inflammation, and a full Systems Age score which “re-unifies” those scores into a single measurement of biological age. Specificity of system scores was assessed using data from a different study. The researchers tested associations with disease incidence, disease prevalence, and various functional parameters of aging.
The tests showed high levels of correlation between system scores and the corresponding aging phenotypes (for instance, between brain score and cognition, heart score and cardiovascular events, and musculoskeletal score and diabetes). Interestingly, the inflammation score was highly correlated with the total burden of comorbidities at baseline, supporting the hypothesis that inflammation has an especially wide and multi-pronged effect on aging.
Comparison to existing clocks
The researchers then compared their system scores to three existing top-tier clocks, GrimAge, PhenoAge, and DunedinPACE, using various aging outcomes. The relevant system scores proved more predictive that the three clocks for 10 out of 14 diseases and came close second in the rest. While the existing clocks were slightly more accurate in some cases, the system scores were more consistent, showing that they are less skewed towards particular aging phenotypes. According to the authors, “these results suggest that having different scores for each system may more precisely capture disease relevant risk and facilitate personalized interventions compared to a single global metric.”
Interestingly, the unified Systems Age score was also placed either first or close second for most of the 14 conditions, while GrimAge and DunedinPACE were overall less consistent. Probably due to that higher consistency, Systems Age also outperformed the other clocks in general categories such as mortality, comorbidities at baseline, and cognitive and physical function.
Some system scores were more highly intercorrelated than others, such as heart and lung along with inflammation and musculoskeletal. Analyzing these correlations, the researchers identified nine clusters (aging subtypes) that showed distinct patterns across the system aging scores and different associations with certain diseases.
Morgan Levine said to Lifespan.io:
Epigenetic clocks exhibit robust age correlations and can be applied universally to almost any tissue or biofluid. However, existing clocks provide only a single value meant to represent the predicted age or rate of aging in the organ or biofluid from which the DNA sample was taken. This is despite us knowing that not every cell, tissue, or even organ in our body is aging at the same rate. To date, the only way to assess differential aging across organ systems using epigenetic clocks has been to take biopsies and then profile each organ individually. To address this, we developed multiple novel systems-based methylation clocks that, when assessed in blood using a single DNA sample and assay, capture aging in distinct physiological systems.
Literature
[1] Sehgal, R., Meer, M., Shadyab, A. H., Casanova, R., Manson, J. E., Bhatti, P., … & Levine, M. (2023). Systems Age: A single blood methylation test to quantify aging heterogeneity across 11 physiological systems. bioRxiv, 2023-07.
[2] Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome biology, 14(10), 1-20.
[3] Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S., … & Horvath, S. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging (albany NY), 10(4), 573.